Dance of the proteins:
Protein motion measured in nanosecond resolution
04.03.2010, Press releases
Researchers at the Department of Chemistry at the Technische Universitaet Muenchen (TUM) have developed a method that allows the observation of local movements in proteins on a time scale of nanoseconds to microseconds. Upon examining motion of the protein villin using this method they found two structures that were otherwise barely distinguishable from one another. Quick nanosecond-scale structure changes essential for the protein function can take place in the one, while the other remains rigid. These results have been published in the current online issue of the journal “Proceedings of the Natural Academy of Sciences, USA” (PNAS).
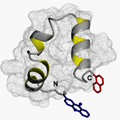
One of the five most important proteins in a cell is actin. Its filaments hold the cell together and keep its key components in their places. The protein villin links the actin filaments and thus contributes significantly to the stabilization of cell structures. Because of its small size, HP35, the part of the villin protein responsible for binding the actin filaments, has been the subject of numerous computer simulations aimed at understanding protein dynamics. However, there have been no experimental studies, as these protein motion takes place on a scale of microseconds and even nanoseconds – a time scale that has, for all intents and purposes, eluded experimental access, until now.
A method developed by Professor Thomas Kiefhaber’s work group, based on fast electron transfer between the different parts of a protein, now makes it possible for the first time to directly examine fast structural changes. They selected the actin-binding part of the villin protein, HP35, as a model system. The new experimental work by Thomas Kiefhaber’s team has shown that the folded protein is available in two conformations that are very similar structurally, but display decidedly different dynamic properties. While significant structural changes are not possible in a rigid conformation, flexible conformations allow parts of the protein responsible for binding actin to fold and unfold on a time scale of 100 nanoseconds.
The two conformations are in equilibrium and transform into one another within one microsecond. The structural similarity of the two conformations explains why they were not previously discovered – neither in structural examinations nor in computer simulations. Using time-resolved electron transfer measurements it is now possible to differentiate between and characterize the different states based on their different motilities.
The insights from this study are fundamental to understanding the function of proteins and will help shed light of the mechanisms behind the folding and misfolding of proteins. The scientists now hope to further develop this method in order to apply it to larger proteins important for the regulation of cell functions.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB 749) and the Cluster of Excellence Center for Integrated Protein Science Munich (CIPSM).
Original publication:
Andreas Reiner, Peter Henklein and Thomas Kiefhaber
An unlocking/relocking barrier in conformational fluctuations of villin headpiece subdomain
Proc. Natl. Acad. Sci. USA, Early Online Edition. doi: 10.1073/pnas.0910001107
Computer simulations on HP35 folding:
http://www.youtube.com/watch?v=AlfvWESPyZY
http://www.youtube.com/watch?v=gvKu3cDeoeA
Contact:
Prof. Dr. Thomas Kiefhaber
Technische Universitaet Muenchen
Chair for Biophysical Chemistry
Lichtenbergstr. 4
85748 Garching, Germany
Tel: +49 89 289 13420 -
Fax: +49 89 289 13416
E-Mail -
Internet
Kontakt: presse@tum.de
More Information
| 100303_HP35_PW.pdf |
Druckversion der Pressemitteilung,
(Type: application/pdf,
Size: 135.2 kB)
Save attachment
|
|
| 100304_hp35_pw.pdf |
Druckversion der Presseinformation (DE),
(Type: application/pdf,
Size: 135.2 kB)
Save attachment
|
|
| 100304_hp35_pw_en.pdf |
Printable version of the press release (EN),
(Type: application/pdf,
Size: 129.9 kB)
Save attachment
|