Bioinformatics strategy helps elucidate protein structures:
How does sugar get into the cell?
08.04.2011, Press releases
Membrane proteins play a significant role in all organisms. They are essential for the communication between cells and their environments and ensure the transport of important substances through the cell membrane. Thanks to their special characteristics these proteins provide an ideal basis for medications. Using a new bioinformatics approach, scientists of the Columbia University, New York and the Technische Universitaet Muenchen (TUM) succeeded in elucidating the structure of an important sugar transporter. The results are presented in the current issue of the journal Nature.
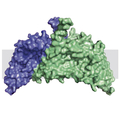
In a broad-ranging cooperation, scientists of Columbia University have succeeded in determining the three-dimensional structure of the bacterial membrane protein ChbC via crystal structure analysis. The protein family that ChbC belongs to is essential for the transport of specific sugars in cells and is, as such, crucial to the survival of the bacterium. However, the precise mechanism by which these proteins channel sugar into the cell was unknown, until now.
Preliminary work of the New York Consortium on Membrane Protein Structure (NYCOMPS) made these results possible. A particular focus of the scientists are membrane proteins. “One in four proteins are membrane proteins and yet we know the structure of only around five percent of them. Here we are still facing a major challenge,” says Burkhard Rost, Professor for Bioinformatics at the TU Muenchen. The pool of potentially interesting sequences is large. The scientists had a pool of over 300,000 protein sequences from the genomes of 96 organisms to choose from. In light of the fact that the experimental analysis of a single membrane protein can take several years, the selection of promising candidates for the time-consuming analysis is of great significance.
Together with other NYCOMPS-bioinformaticians Burkhard Rost identified ChbC as a promising candidate. Their starting point is a bioinformatics method from the field of “structural genomics.” The basic assumption of this method is that proteins with common evolutionary anscestors, so-called “protein families,” bear similarities in both their amino acid sequences and their three-dimensional structure. Once the structure of one of the related proteins is known, those of the the others can be predicted.
In an attempt to find a suitable protein, the NYCOMPS reserachers refined a standard bioinformatics approach. Instead of following the standard procedure of ordering all proteins in a comprehensive genome reference map to then make a selection, the bioinformaticians ordered each of the protein families along a single sequence. The advantage of such “seed proteins” is that the scientists can immediately expand interesting target sequences into protein families, instead of first having to search for them in the protein reference maps. The likelihood of finding a relative suitable for the ensuing experiments within the family created about the target protein is also higher.
The NYCOMPS scientists filtered out proteins that showed charateristics of membrane proteins, one by one. Then they expanded these into protein families and slected those sequences that were particularly suited for the experiments. All of this happened virtually, using mathematical predictions in lieu of experiments. In the case of ChbC, in the end only 25 sequences from 13 organisms remained out of the multitude of original sequences. Only these 25 were then closely scrutinized in the laboratory. Ultimately, one of the sequences led to success – the proverbial “needle in the haystack” had beed found.
The newly deciphered structure of ChbC is now helping scientists better understand a key mechanism of sugar transport into the cells: the so-called “phosphotransferase system” (PTS). In this transport mechanism the sugar molecule is modified with a phosphate residue during its transport into the cell. This modification is the first step in the later process of extracting energy from the sugar, while at the same time preventing the sugar from simply exiting the cell again. Phototransferase systems appear in many kinds of bacteria and influence a wide variety of metabolic pathways in cells. As such, they serve as promising targets for antibiotics.
The structure of ChbC is exemplary for how such transport systems might function. The part of ChbC that attaches to the sugar molecule to be transported is similar in structure to another transporter that is not related to ChbC, but whose transport mechanism had been previously researched: the protein GltPh. The idea was that the transport mechanism of ChbC might function in a manner simmilar to GltPh.
Prior to this, the precise transport mechanism of phosphotransferase systems was unknown. Now, the scientiscts suspect that the sugar transport in ChbC takes place in three phases. In the first, “open to the outside” state, the protein binds to the sugar to be transported. The region the sugar is attached to flips into an interim state closer to the inside wall of the cell membrane, without yet having contact with the inside of the cell. In the last stage, the modified sugar is finally released into the inside of the cell. A single transport domain that moves relative to the remaining stationary part of the cell may be responsible for the movement between states.
This work was supported by the US National Institutes of Health, the NYCOMPS central facility is part of the Protein Structure Initiative (PSI-2) established by the National Institute of General Medical Sciences. Burkhard Rost is Alexander von Humboldt professor and Member of the Institute for Advanced Study at the Technische Universitaet Muenchen.
Publication:
Crystal structure of a phosphorylation-coupled saccharide transporter Yu Cao, Xiangshu Jin, Elena J. Levin, Hua Huang, Yinong Zong, Matthias Quick, Jun Weng, Yaping Pan, James Love, Marco Punta, Burkhard Rost, Wayne A. Hendrickson, Jonathan A. Javitch, Kanagalaghatta R. Rajashankar, Ming Zhou Nature, publishd online, 6 April 2011
http://www.nature.com/nature/journal/vaop/ncurrent/full/nature09939.html?WT.ec_id=NATURE-20110407
Kontakt: presse@tum.de
More Information
| 110408_ChbC-Rost_DE.pdf |
Druckversion der Presseinformation (DE),
(Type: application/x-pdf,
Size: 105.7 kB)
Save attachment
|
|
| 110408_ChbC-Rost_EN.pdf |
Printable version of the press release (EN),
(Type: application/x-pdf,
Size: 94.7 kB)
Save attachment
|